Tn6019
Author: Nabil Karah and Bernt Eric Uhlin
Department of Molecular Biology, Umeå University, Umeå, Sweden (nabil.karah@umu.se; bernt.eric.uhlin@umu.se)
Reviewer: Ruth M Hall
School of Life and Environmental Sciences, The University of Sydney, Sydney, Australia (ruth.hall@sydney.edu.au)
Genetic structure of Tn6019
Tn6019 was first described as the backbone of AbaR5, a resistance island inserted in the comM gene of the chromosome of Acinetobacter baumannii strain 3208 [Australia, 1997, sequence type 1 according to the Pasteur multilocus sequence typing scheme (ST1P), global clone (GC) 1] (Post & Hall, 2009; Post et al., 2010).
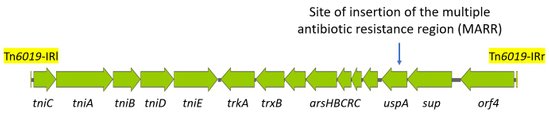
Tn6019 is 16332 base pair (bp) long (Figure 1, Hamidian & Hall, 2018). It is bounded by two imperfect inverted repeats (IRs) of 26 bp, sharing 19/26 identity to the reverse complement sequence of each other, namely: IR1: TGTCATTTACAGCAATAGAATAGAGT and IR2: ACTCTGATTTATTTGTGGAACTGACA.
Tn6019 consists of 15 open reading frames (orf), among which the first 5 orfs correspond to genes (tniC, tniA, tniB, tniD, and tniE) with potential transposition function (Table 1 & Figure 1). An operon of five genes, arsHBCRC, conferring resistance to arsenite and arsenate (Hamidian & Hall, 2018), is also present. Other genes include trkA (putative monooxygenase), trxB (thioredoxin-disulfide reductase), uspA (universal stress protein), and sup (sulphate permease). Only the last orf has no predicted function and is usually annotated as orf4.
Figure 1. Genetic structure of Tn6019 (adapted from Post et al., 2010 and Hamidian & Hall, 2018)
AbaR0- and AbaR3-type resistance islands
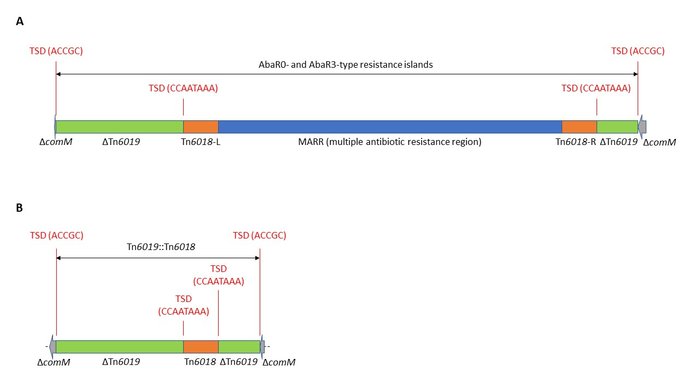
The uspA gene of Tn6019 in AbaR5 was interrupted by a multiple antibiotic resistance region (MARR) flanked by two copies of Tn6018 (Figure 2a, FJ172370.5, Post & Hall, 2009). The insertion of Tn6019 (AbaR5) had generated a characteristic target site duplication of 5 bp (ACCGC).
Tn6019 is the common backbone of all the AbaR0- and AbaR3-type resistance islands (Post et al., 2010; Hamidian & Hall, 2018) that have the same general structure as AbaR5. AbaR0 (GenBank accession no.: KF483599.2), first detected in the chromosome of A. baumannii WM98 (Australia, 1998, GC1), is the most complete form of these AbaR islands. It was proposed as the early ancestor of the AbaR3-type resistance islands (Hamidian et al., 2014). These AbaR islands and their many deletion derivatives are found in the comM gene in the chromosomes of lineage 1 of GC1 (Holt et al., 2016).
Tn6019::Tn6018
Tn6019 with uspA interrupted by only one copy of Tn6018(Figure 2b), was detected in the complete genome of the GC1 lineage 1 A. baumannii strain AB5075-UW (USA, 2008, ST1P, CP008706.1) (Jacobs et al., 2014; Gallagher et al., 2015). It is also inserted in comM and likely arose from an AbaR via homologous recombination between the two Tn6018 copies. Draft genome records of few other isolates in GenBank (Table 2) were also found to include Tn6019::Tn6018. However, the presence of this configuration in draft assemblies could be due to an assembly error and further data would be needed to confirm that the MARR is missing. Our preliminary analysis shows that many of the Tn6019::Tn6018-positive isolates belong to ST1P.
Figure 2. Genetic structures of (a) AbaR5 as a representative of the AbaR0- and AbaR3-type resistance islands (adapted from Post & Hall, 2009), and (b) Tn6019::Tn6018 in A. baumannii AB5075-UW.
Tn6019 and Tn6019::ISAba1
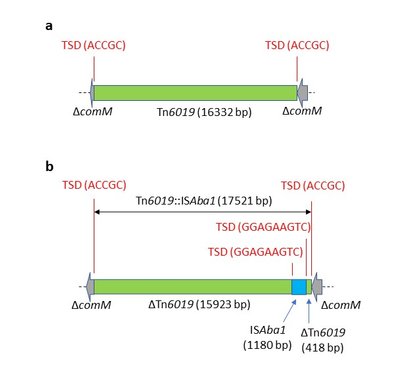
A full-length version of Tn6019 that is not interrupted by a MARR or by any insertion sequence (IS) element was detected in the comM gene in the genome records of few A. baumannii isolates, such as MRSN31942 (VHFO01000032.1; Figure 3a), MRSN7251 (VHED01000094.1), 4300STDY7045780 (UFLD01000011.1), TUM15181 (BKJY01000003.1), and NCTC 13305 (UFSC01000003.1). Tn6019 was also detected in the genomic record of A. baumannii ARLG1306 (NGHM01000051.1), though it was inserted in a different chromosomal site (in an orf encoding a putative lipoprotein). The intact version of Tn6019 was plasmid associated in the genomic records of A. baumannii 2019KU-00110 (ABDOFH010000008.1) and 2020JQ-00122 (ABDUQV010000016.1), though the two plasmids were different.
Tn6019 with orf4 interrupted by ISAba1 (Tn6019::ISAba1) was detected in the genomic records of few other isolates in GenBank, such as A. baumannii strains AR_0052 (CP027183.1; Figure 3b), AR_0070 (CP027178.1), and probably in ARLG-1301 (NGER01000041.1 and NGER01000118.1). Interestingly, most of the A. baumannii isolates with Tn6019 or Tn6019::ISAba1 belong to ST32P (1, 1, 2, 2, 3, 4, 4) or its single locus variant ST128P (1, 38, 2, 2, 3, 4, 4) as shown in Table 3. Exceptions are isolates 2019KU-00110 which is ST2P (2, 2, 2, 2, 2, 2, 2) and 2020JQ-00122 which belongs to ST734P (1, 1, 1, 1, 3, 1, 1). However, in both of these the Tn could be plasmid associated.
Figure 3. Genetic structures of (a) Tn6019 in A. baumannii MRSN31942, and (b) Tn6019::ISAba1 in A. baumannii AR_0052
Tn6019::Tn7587
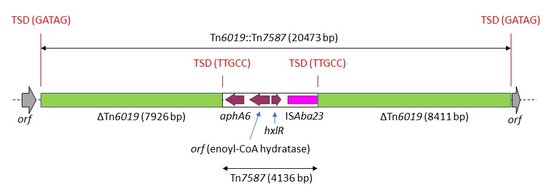
Detailed examination of the genomic record for plasmid p1OC041 (CP087358.1) found in A. baumannii isolate OC041 enabled us to detect a novel transposon (4136 bp) inserted in the trxB gene of Tn6019 (Figure 4). The novel transposon, designated Tn7587 (number was allocated by TheTransposon Registry, https://transposon.lstmed.ac.uk/), had a single copy of ISAba23 (1249 bp) on one side. The first 20-bp sequence (AAAATCTTTGCACGATGAAA) of Tn7587 had significant nucleotide similarity with part of the left inverted repeat of ISAba23 (GAAACCTTTGCACGAATAAA), which could indicate that this 20-bpwere used as an alternative IRL for ISAba23 (as previously suggested for Tn2008, Wang etal., 2011). Tn7587 carried 3 orfs, including the amikacin resistance gene aphA6, and was probably acquired from the chromosome of Acinetobacter guillouiae (Yoon et al., 2014). The aphA6 gene in Tn7587 has 757/780 (97%) nucleotide identity to the commonly reported version of aphA6 in A. baumannii (X07753.1; JF343537.1) (Martin et al., 1988; Nigro et al., 2011). Insertionof Tn7587 created a 5-bp target site duplication. Tn6019::Tn7587 was inserted between two orfs with no putative conserved domains.
Figure 4. Genetic structure of Tn6019::Tn7587 in A. baumannii OC041
Tn6019 in other Acinetobacter species
Interestingly, Acinetobacter lwoffii isolate WU_MDCI_Al101 (AHPSO010000005.1) and Acinetobacter radioresistens isolates C21A (JAMOZF010000010.1) and C30P (JAMOZE010000012.1) also carried an intact version of Tn6019. A related version showing 93% nucleotide identity to Tn6019 was also detected in the genomic record of Acinetobacter shaoyimingii isolate 323-1 (CP049801.1). Tn6019 was inserted in the chromosomes of the latter four non-baumannii isolates in genes showing around 70% nucleotide identities to the comM gene of A. baumannii.
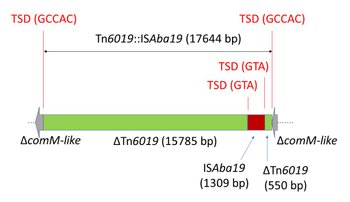
Tn6019 with orf4 interrupted by ISAba19 (Tn6019::ISAba19) was detected in the genomic records of Acinetobacter haemolyticus isolate AN54 (CP041224.1; Figure 5) and probably in A. haemolyticus 5439 (WTTV01000022.1 and WTTV01000098.1). Tn6019::ISAba19 was inserted in chromosomal genes showing 71% nucleotide identity to the comM gene of A. baumannii.
Figure 5. Genetic structure of Tn6019::ISAba19 in A. haemolyticus AN54
References
Post V, Hall RM. AbaR5, a large multiple-antibiotic resistance region found in Acinetobacter baumannii. Antimicrob Agents Chemother. 2009 Jun;53(6):2667-71.
DOI: 10.1128/AAC.01407-08
PMID: 19364869
Post V, White PA, Hall RM. Evolution of AbaR-type genomic resistance islands in multiply antibiotic-resistant Acinetobacter baumannii. J Antimicrob Chemother. 2010 Jun;65(6):1162-70.
DOI: 10.1093/jac/dkq095
PMID: 20375036
Hamidian M, Hall RM. The AbaR antibiotic resistance islands found in Acinetobacter baumannii global clone 1 - Structure, origin and evolution. Drug Resist Updat. 2018 Nov;41:26-39.
DOI: 10.1016/j.drup.2018.10.003
PMID: 30472242
Hamidian M, Wynn M, Holt KE, Pickard D, Dougan G, Hall RM. Identification of a marker for two lineages within the GC1 clone of Acinetobacter baumannii. J Antimicrob Chemother. 2014 Feb;69(2):557-8.
DOI: 10.1093/jac/dkt379
PMID: 24080502
Holt K, Kenyon JJ, Hamidian M, Schultz MB, Pickard DJ, Dougan G, Hall R. Five decades of genome evolution in the globally distributed, extensively antibiotic-resistant Acinetobacter baumannii global clone 1. Microb Genom. 2016 Feb 23;2(2):e000052.
Erratum in: Microb Genom. 2019 Jul;5(7)
PMID: 28348844
Jacobs AC, Thompson MG, Black CC, Kessler JL, Clark LP, McQueary CN, Gancz HY, Corey BW, Moon JK, Si Y, Owen MT, Hallock JD, Kwak YI, Summers A, Li CZ, Rasko DA, Penwell WF, Honnold CL, Wise MC, Waterman PE, Lesho EP, Stewart RL, Actis LA, Palys TJ, Craft DW, Zurawski DV. AB5075, a highly virulent isolate of Acinetobacter baumannii, as a model strain for the evaluation of pathogenesis and antimicrobial treatments. mBio. 2014 May 27;5(3):e01076-14.
PMID: 24865555
Gallagher LA, Ramage E, Weiss EJ, Radey M, Hayden HS, Held KG, Huse HK, Zurawski DV, Brittnacher MJ, Manoil C. Resources for genetic and genomic analysis of emerging pathogen Acinetobacter baumannii. J Bacteriol. 2015 Jun 15;197(12):2027-35.
DOI: 10.1128/JB.00131-15
PMID: 25845845
Wang X, Zong Z, Lü X. Tn2008 is a major vehicle carrying blaOXA-23 in Acinetobacter baumannii from China. Diagn Microbiol Infect Dis. 2011 Feb;69(2):218-22.
DOI: 10.1016/j.diagmicrobio.2010.10.018
PMID: 21251570
Yoon EJ, Goussard S, Touchon M, Krizova L, Cerqueira G, Murphy C, Lambert T, Grillot-Courvalin C, Nemec A, Courvalin P. Origin in Acinetobacter guillouiae and dissemination of the aminoglycoside-modifying enzyme Aph(3')-VI. mBio. 2014 Oct 21;5(5):e01972-14.
PMID: 25336457
Martin P, Jullien E, Courvalin P. Nucleotide sequence of Acinetobacter baumannii aphA-6 gene: evolutionary and functional implications of sequence homologies with nucleotide-binding proteins, kinases and other aminoglycoside-modifying enzymes. Mol Microbiol. 1988 Sep;2(5):615-25.
DOI: 10.1111/j.1365-2958.1988.tb00070.x
PMID: 2846986
Nigro SJ, Post V, Hall RM. Aminoglycoside resistance in multiply antibiotic-resistant Acinetobacter baumannii belonging to global clone 2 from Australian hospitals. J Antimicrob Chemother. 2011 Jul;66(7):1504-9.
DOI: 10.1093/jac/dkr163
PMID: 21586593