oxaAb (blaOXA-51-like)
Author: Benjamin A Evans
Norwich Medical School, University of East Anglia, Norwich, United Kingdom (Benjamin.Evans@uea.ac.uk)
Reviewer: Andrés Opazo-Capurro
Laboratorio de Investigación en Agentes Antibacterianos (LIAA) - Universidad de Concepción, Concepción, Chile (andopazo@udec.cl)
oxaAb are intrinsic to A. baumannii
OXA-type beta-lactamases are found in many bacterial species and comprise the largest family of beta-lactamases (Yoon and Jeong, 2021). It is thought that all members of the Acinetobacter genus carry the gene for an OXA-type beta-lactamase on their chromosome. More specifically, each Acinetobacter species has a distinct OXA-type beta-lactamase that is characteristic of the species. In the case of A. baumannii, this is the OxaAb (or OXA-51-like) beta-lactamase.
Nomenclature
The nomenclature of OXA-type beta-lactamases, which belong to the class D beta-lactamases, is based upon the principle that each different amino-acid sequence is given a new allele number in a purely sequential manner, irrespective of whether these alleles derive from the same gene or not. The first intrinsic OXA beta-lactamase gene identified in A. baumannii was therefore named blaOXA-51 following this naming convention (Brown, Young and Amyes, 2005). As the number of different alleles for this gene increased, the term blaOXA-51-like was adopted to describe the group of alleles.
Some authors have recently started referring to the OXA-51-like enzymes as the OxaAb enzymes, and the blaOXA-51-like genes as oxaAb genes, with the ‘Ab’ signifying that these are the OXA beta-lactamases intrinsic to A. baumannii (Nigro and Hall, 2016; Nigro and Hall, 2018; Takebayashi et al., 2021). The allele for the gene is then given in parenthesis e.g. oxaAb(51) is the same as blaOXA-51. It is proposed that this system preserves the information regarding the specific allele while simultaneously identifying the specific gene the alleles belong to. This allows easy distinction between these alleles and those belonging to other OXA beta-lactamases found both various Acinetobacter species and in other genera. Both of these nomenclatures are currently used in the literature.
The number of oxaAb alleles that have been identified has rapidly increased, and currently numbers more than 370 alleles. These can be viewed in the NCBI Bacterial Antimicrobial Resistance Reference Gene Database (Reference Gene Catalog - Pathogen Detection - NCBI (nih.gov)).
Association with epidemic lineages
As the oxaAb gene is an intrinsic chromosomal gene in A. baumannii, much of the allelic diversity reflects the population structure of the species, with different alleles being representative of different bacterial lineages. A clear example of this is found with the international clones (ICs), also referred to as global clones (GCs), where a specific oxaAb allele is associated with the clone (Table 1).
Table 1: Association between international clonal lineages of A. baumannii and specific oxaAb alleles.
Clone | oxaAb / blaOXA-51-like allele | References |
IC1 | oxaAb(69) / blaOXA-69 | |
IC2 | oxaAb(66) / blaOXA-66 | |
IC3 | oxaAb(71) / blaOXA-71 | |
IC4 | oxaAb(51) / blaOXA-51 | |
IC5 | oxaAb(65) / blaOXA-65 | |
IC6 | oxaAb(90) / blaOXA-90 | |
IC7 | oxaAb(64) / blaOXA-64 | |
IC8 | oxaAb(68) / blaOXA-68 | |
IC9 | oxaAb(94) / blaOXA-94 |
Role in antimicrobial resistance
A small number of OxaAb enzymes have been purified and their kinetics measured to determine the ability to hydrolyse beta-lactam antibiotics. OxaAb enzymes are able to hydrolyse penicillins but generally not cephalosporins. While it is thought most OxaAb variants are not particularly effective at hydrolysing clinically used carbapenem antibiotics, a few variants with amino acid substitutions at particular sites (e.g. positions 129, 130, 167, 222) appear to be able to hydrolyse carbapenems, and in one instance, late generation cephalosporins (Brown, Young and Amyes, 2005; Héritier et al., 2005; Zander et al., 2013; Mitchell and Leonard, 2014; Schroder et al., 2016; Takebayashi et al., 2021; Chan et al., 2022).
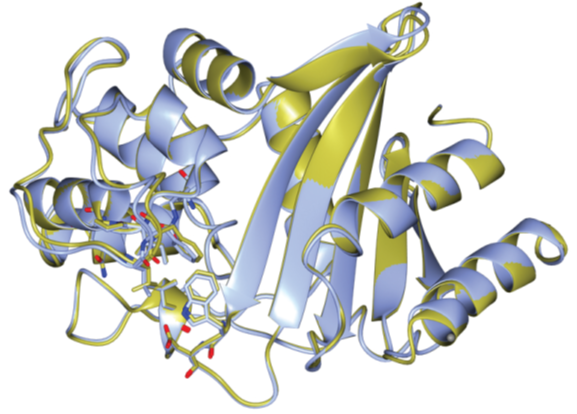
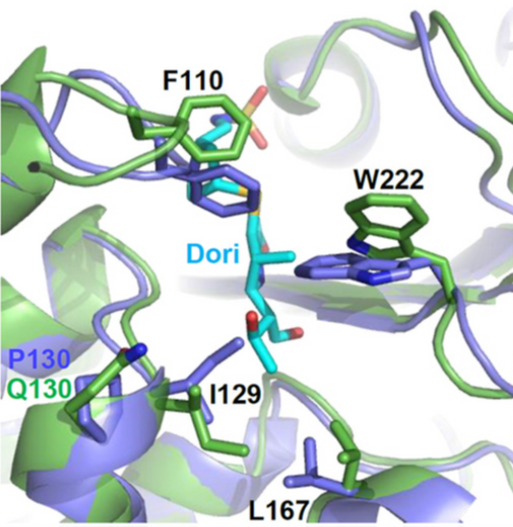
The crystal structures of OxaAb(51) and OxaAb(66) have been solved (Figure 1) (Smith et al., 2015; June et al., 2016; Takebayashi et al., 2022). Analyses of the amino acid positions listed above confirmed their involvement in altering the activity of the enzyme through altering the environment of the active site and facilitating greater access for the antibiotics (Figure 2).
Figure 1: Comparison of the structures of OxaAb(66) (blue) and OxaAb(51) (gold) (Takebayashi et al., 2022).
Figure 2: Comparison of the structures of OxaAb(66) (blue) and OxaAb(109) (green) with the carbapenem doripenem (cyan) in the active site region, showing alternative conformations [Adapted from Schroder et al., (2016)].
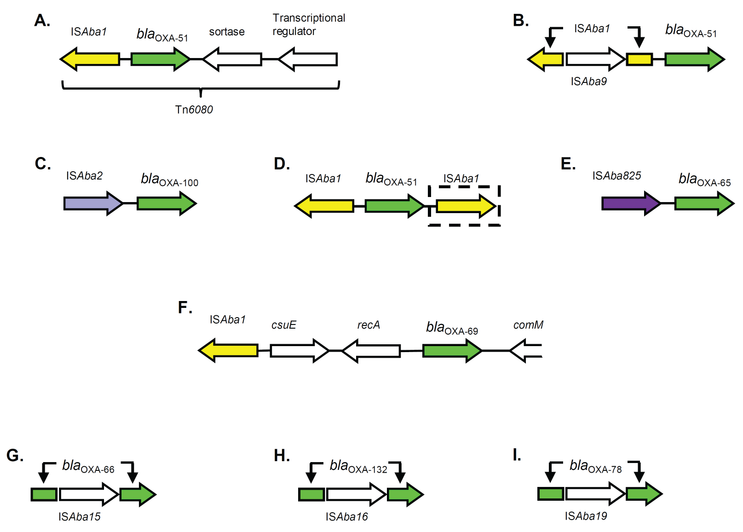
A. baumannii is able to drive increased expression levels of the oxaAb genes through the integration of insertion sequences (IS) upstream of the oxaAb gene. Here, the IS provides a strong promotor that drives increased expression of this gene (Figure 3) (Turton et al., 2006; Figueiredo et al., 2009a). By far the most common IS that does this is ISAba1. In a few rare instances, IS elements have been found to insert into the oxaAb gene, inactivating it.
It has been noted that the frequency of oxaAb alleles that can be found with an IS element upstream of them is not equal across all alleles. The alleles that encode OxaAb variants with mutations at amino acid positions that increase the carbapenemase activity of the enzymes as described above are more often found to have an ISAba1 element upstream than other alleles that do not effectively hydrolyse carbapenems (Evans et al., 2008).
Figure 3: Transposons associated with oxaAb genes. A, (Chen et al., 2010); B, (Figueiredo et al., 2009b); C, (Al-Hassan, El Mehallawy and Amyes, 2013); D, (Turton et al., 2006; Espinal et al., 2009); E, (Lopes, Al-Hassan and Amyes, 2012); F, (Post, Hamidian and Hall, 2012); G, (Zander et al., 2013b); H, (Lopes, Evans and Amyes, 2012); I, (Zander et al., 2013b). Adapted from (Evans and Amyes, 2014).
Evolution of the oxaAb genes
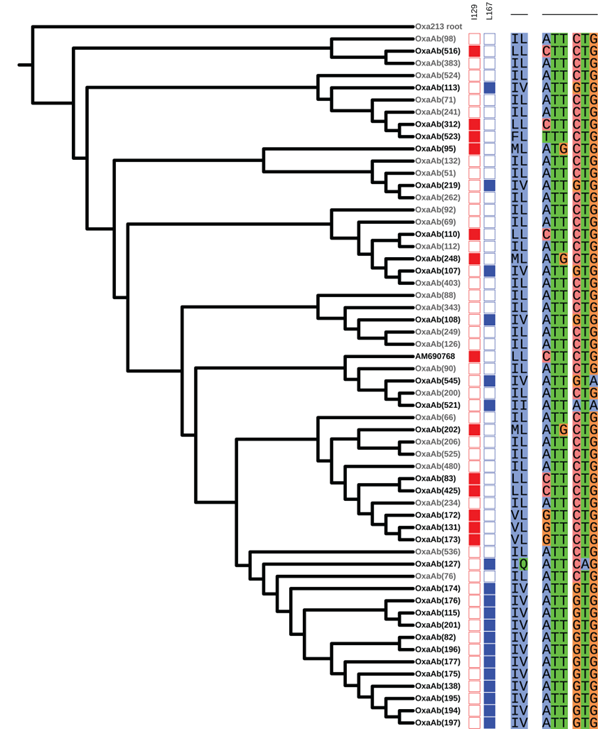
While much of the diversity observed in oxaAb alleles likely reflects the underlying genetic diversity of the species, there is evidence that certain alleles have arisen due to antibiotic selective pressure. Enzyme variants with differences at amino acid sites known to be responsible for increasing the ability of the enzymes to hydrolyse carbapenems appear to have been selected for on multiple occasions as they arise in many separate enzyme variants across the phylogenetic tree (Figure 4).
Figure 4: Phylogeny of selected OxaAb proteins (Takebayashi et al., 2021). Variants with a change from consensus at positions 129 or 167 are shown in bold text. Filled coloured boxes indicate the enzyme has a change at the indicated position. To the right of the coloured boxes are shown the amino acids at the two positions, and on the far right the codons at the two positions.
Mobilised oxaAb genes
While the oxaAb genes are normally chromosomally located, there have been rare reports of the gene being located on a plasmid. To date these have been from Taiwan. Plasmids carrying oxaAb were found in Acinetobacter nosocomialis and Acinetobacter seifertii (Lee et al., 2009; Chen et al., 2010; Lee et al., 2012; Li et al., 2021). The oxaAb gene appears to have been mobilised onto plasmids via ISAba1-mediated single-ended transposition. The reason why these plasmids have only been observed in Taiwan to date is unclear. Unlike some other OXA-type enzymes such as Oxa23, it is very rare for oxaAb genes to be located in a composite transposon, which may have contributed to their limited spread outside of A. baumannii.
References
Yoon EJ, Jeong SH. Class D Beta-lactamases. J Antimicrob Chemother. 2021 Mar;76(4): 836-864.
doi: 10.1093/jac/dkaa513
PMID: 33382875
Brown S, Young HK, Amyes SG. Characterisation of OXA-51, a novel class D carbapenemase found in genetically unrelated clinical strains of Acinetobacter baumannii from Argentina. Clin Microbiol Infect. 2005 Jan;11(1):15-23.
doi: 10.1111/j.1469-0691.2004.01016.x
PMID: 15649299
Nigro SJ, Hall RM. Structure and context of Acinetobacter transposons carrying the oxa23 carbapenemase gene. J Antimicrob Chemother. 2016 May;71(5):1135-47.
doi: 10.1093/jac/dkv440
PMID: 26755496
Nigro SJ, Hall RM. Does the intrinsic oxaAb (blaOXA-51-like) gene of Acinetobacter baumannii confer resistance to carbapenems when activated by ISAba1? J Antimicrob Chemother. 2018 Dec 1;73(12):3518-3520.
doi: 10.1093/jac/dky334
PMID: 30124881
Takebayashi Y, Findlay J, Heesom KJ, Warburton PJ, Avison MB, Evans BA. Variability in carbapenemase activity of intrinsic OxaAb (OXA-51-like) β-lactamase enzymes in Acinetobacter baumannii. J Antimicrob Chemother. 2021 Feb 11;76(3):587-595.
doi: 10.1093/jac/dkaa502
PMID: 33338207
Turton JF, Gabriel SN, Valderrey C, Kaufmann ME, Pitt TL. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin Microbiol Infect. 2007 Aug;13(8):807-15.
doi: 10.1111/j.1469-0691.2007.01759.x
PMID: 17610600
Evans BA, Hamouda A, Towner KJ, Amyes SG. OXA-51-like beta-lactamases and their association with particular epidemic lineages of Acinetobacter baumannii. Clin Microbiol Infect. 2008 Mar;14(3):268-75.
doi: 10.1111/j.1469-0691.2007.01919.x
PMID: 18190566
Zander E, Nemec A, Seifert H, Higgins PG. Association between β-lactamase-encoding bla(OXA-51) variants and DiversiLab rep-PCR-based typing of Acinetobacter baumannii isolates. J Clin Microbiol. 2012 Jun;50(6):1900-4.
doi: 10.1128/JCM.06462-11
PMID: 22422849
Al-Hassan L, Elbadawi H, Osman E, Ali S, Elhag K, Cantillon D, Wille J, Seifert H, Higgins PG. Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii from Khartoum state, Sudan. Front Microbiol. 2021 Feb 26;12:628736. doi: 10.3389/fmicb.2021.628736
PMID: 33717019
Héritier C, Poirel L, Fournier PE, Claverie JM, Raoult D, Nordmann P. Characterization of the naturally occurring oxacillinase of Acinetobacter baumannii. Antimicrob Agents Chemother. 2005 Oct;49(10):4174-9.
doi: 10.1128/AAC.49.10.4174-4179.2005
PMID: 16189095
Mitchell JM, Leonard DA. Common clinical substitutions enhance the carbapenemase activity of OXA-51-like class D β-lactamases from Acinetobacter spp. Antimicrob Agents Chemother. 2014 Nov;58(11):7015-6.
doi: 10.1128/AAC.03651-14
PMID: 25155607
Zander E, Chmielarczyk A, Heczko P, Seifert H, Higgins PG. Conversion of OXA-66 into OXA-82 in clinical Acinetobacter baumannii isolates and association with altered carbapenem susceptibility. J Antimicrob Chemother. 2013 Feb;68(2):308-11.
doi: 10.1093/jac/dks382
PMID: 23014718
Schroder EC, Klamer ZL, Saral A, Sugg KA, June CM, Wymore T, Szarecka A, Leonard DA. Clinical variants of the native Class D β-Lactamase of Acinetobacter baumannii pose an emerging threat through increased hydrolytic activity against carbapenems. Antimicrob Agents Chemother. 2016 Sep 23;60(10):6155-64.
doi: 10.1128/AAC.01277-16
PMID: 27480863
Chan KW, Liu CY, Wong HY, Chan WC, Wong KY, Chen S. Specific amino acid substitutions in OXA-51-type β-lactamase enhance catalytic activity to a level comparable to carbapenemase OXA-23 and OXA-24/40. Int J Mol Sci. 2022 Apr 19;23(9):4496.
doi: 10.3390/ijms23094496
PMID: 35562886
Smith CA, Antunes NT, Stewart NK, Frase H, Toth M, Kantardjieff KA, Vakulenko S. Structural basis for enhancement of carbapenemase activity in the OXA-51 family of class D β-lactamases. ACS Chem Biol. 2015 Aug 21;10(8):1791-6.
doi: 10.1021/acschembio.5b00090
PMID: 26042471
June CM, Muckenthaler TJ, Schroder EC, Klamer ZL, Wawrzak Z, Powers RA, Szarecka A, Leonard DA. The structure of a doripenem-bound OXA-51 class D β-lactamase variant with enhanced carbapenemase activity. Protein Sci. 2016 Dec;25(12):2152-2163.
doi: 10.1002/pro.3040
PMID: 27636561
Takebayashi Y, Henderson SR, Chirgadze DY, Warburton PJ, Evans BA. OXA-66 structure and oligomerisation of OXAAb enzymes. Access Microbiol. 2022 Oct 3;4(10):acmi000412.
PMID: 36415731
Turton JF, Ward ME, Woodford N, Kaufmann ME, Pike R, Livermore DM, Pitt TL. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol Lett. 2006 May;258(1):72-7.
doi: 10.1111/j.1574-6968.2006.00195.x
PMID: 16630258
Figueiredo S, Poirel L, Croize J, Recule C, Nordmann P. In vivo selection of reduced susceptibility to carbapenems in Acinetobacter baumannii related to ISAba1-mediated overexpression of the natural blaOXA-66 oxacillinase gene. Antimicrob Agents Chemother. 2009 Jun;53(6):2657-9.
doi: 10.1128/AAC.01663-08
PMID: 19307373
Figueiredo S, Poirel L, Papa A, Koulourida V, Nordmann P. Overexpression of the naturally occurring blaOXA-51 gene in Acinetobacter baumannii mediated by novel insertion sequence ISAba9. Antimicrob Agents Chemother. 2009 Sep;53(9):4045-7.
doi: 10.1128/AAC.00292-09
PMID: 19564367
Chen TL, Lee YT, Kuo SC, Hsueh PR, Chang FY, Siu LK, Ko WC, Fung CP. Emergence and distribution of plasmids bearing the blaOXA-51-like gene with an upstream ISAba1 in carbapenem-resistant Acinetobacter baumannii isolates in Taiwan. Antimicrob Agents Chemother. 2010 Nov;54(11):4575-81.
doi: 10.1128/AAC.00764-10
PMID: 20713680
Al-Hassan L, El Mehallawy H, Amyes SG. Diversity in Acinetobacter baumannii isolates from paediatric cancer patients in Egypt. Clin Microbiol Infect. 2013 Nov;19(11):1082-8.
PMID: 23413888
Lopes BS, Al-Hassan L, Amyes SG. ISAba825 controls the expression of the chromosomal blaOXA-51-like and the plasmid borne blaOXA-58 gene in clinical isolates of Acinetobacter baumannii isolated from the USA. Clin Microbiol Infect. 2012 Nov;18(11):E446-51.
doi: 10.1111/j.1469-0691.2012.03979.x
PMID: 22862829
Post V, Hamidian M, Hall RM. Antibiotic-resistant Acinetobacter baumannii variants belonging to global clone 1. J Antimicrob Chemother. 2012 Apr;67(4):1039-40.
doi: 10.1093/jac/dkr586
PMID: 22279182
Zander E, Higgins PG, Fernández-González A, Seifert H. Detection of intrinsic blaOXA-51-like by multiplex PCR on its own is not reliable for the identification of Acinetobacter baumannii. Int J Med Microbiol. 2013 Mar;303(2):88-9.
doi: 10.1016/j.ijmm.2012.12.007
PMID: 23375845
Evans BA, Amyes SG. OXA β-lactamases. Clin Microbiol Rev. 2014 Apr;27(2):241-63.
doi: 10.1128/CMR.00117-13
PMID: 24696435
Lee YT, Kuo SC, Chiang MC, Yang SP, Chen CP, Chen TL, Fung CP. Emergence of carbapenem-resistant non-baumannii species of Acinetobacter harboring a blaOXA-51-like gene that is intrinsic to A. baumannii. Antimicrob Agents Chemother. 2012; 56(2): 1124-7.
doi: https://doi.org/10.1128/aac.00622-11
PMID: 22083478
Lee YT, Turton JF, Chen TL, Wu RC, Chang WC, Fung CP, Chen CP, Cho WL, Huang LY, Siu LK. (2009) First identification of blaOXA-51-like in non-baumannii Acinetobacter spp. J Chemother. 2009; 21(5): 514-20.
doi: 10.1179/joc.2009.21.5.514
PMID: 19933042
Li LH, Yang YS, Sun JR, Huang TW, Huang WC, Chen FJ, Wang YC, Kuo TH, Kuo SC, Chen TL, Lee YT, ACTION Study Group. Clinical and molecular characterization of Acinetobacter seifertii in Taiwan. J Antimicrob Chemother. 2021; 76(2): 312-321.
doi: 10.1093/jac/dkaa432
PMID: 33128052