Horizontal Gene Transfer and Natural Transformation
Author: Sara Domingues
Faculty of Pharmacy, University of Coimbra, Coimbra, Portugal (saradomingues@ff.uc.pt)
Reviewer: Gottfried Wilharm
Robert Koch Institute, Project group P2, Wernigerode, Germany (WilharmG@rki.de)
Horizontal gene transfer
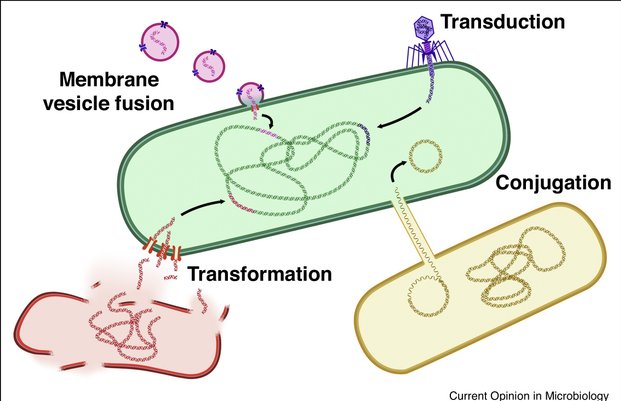
Horizontal gene transfer (HGT) allows the transfer of genetic material between bacteria belonging to the same generation (that it is not offspring), representing a main driver for bacterial adaptation and evolution (Averhoff et al., 2021). HGT mechanisms are particularly important in the context of antimicrobial resistance dissemination. Conjugation, transduction, and natural transformation are the main known HGT mechanisms and a fourth mechanism, vesicle-mediated transfer, has been recently recognized (Figure 1). All these four mechanisms of HGT have been reported in Acinetobacter baumannii, including in the context of antimicrobial resistance dissemination (Da Silva & Domingues, 2016). Despite conjugation is assumed to be the most common HGT mechanism among bacteria, it does not seem the major driving force in acquisition of exogenous DNA in this species, as most sequenced plasmids lack transfer and mobilization genes (Fondi et al., 2010).
Figure 1 - Horizontal gene transfer mechanisms (McInnes et al., 2020, Curr Opin Microbiol).
Natural transformation
Natural transformation is known to occur in the Acinetobacter genus for a long time. In 2010, natural competence was described for the first in A. baumannii (Ramirez et al., 2010). Recently, natural transformation was suggested as the main HGT mechanism involved in antibiotic resistance dissemination in A. baumannii (Averhoff et al., 2021; Godeux et al., 2022). Different studies have determined the conditions that affect transformability of A. baumannii, but most results seem to be strain dependent. A. baumannii is able to uptake both plasmid and chromosomal DNA (Wilharm et al., 2013; Traglia et al., 2016; Godeux et al., 2018). A recent study shows that A. baumannii can efficiently capture long DNA fragments secreted by living cells (Godeux et al., 2022).
Transformation can occur at different pH levels (Traglia et al., 2016; Godeux et al., 2018) and competence can be induced by the presence of calcium ions (Traglia et al., 2016) and sub-inhibitory concentrations of some antibiotics (Quinn et al., 2018). Presence of albumin in the induction of competence seems contradictory, with studies showing both the increase (Traglia et al., 2016; Martinez et al., 2018) and decrease (Godeux et al., 2018) of transformation. Osmolarity has also shown to not affect (Traglia et al., 2016) or to influence (Domingues et al., 2019; Hu et al., 2019) transformability. Natural transformation is less efficient in liquid medium (Traglia et al., 2016) than in semisolid (Domingues et al., 2019) and solid medium (Godeux et al., 2018).
Type IV pili are essential to transformation in A. baumannii (Wilharm et al.,2013; Vesel and Blokesch, 2021), with piliation and transformability peaking at the exponential growth phase (Vesel and Blokesch, 2021). Several genes involved in natural transformation of A. baumannii have been recently identified (Vesel and Blokesch, 2021; Hu et al., 2022).
References
Averhoff B, Kirchner L, Pfefferle K, Yaman D. Natural transformation in Gram-negative bacteria thriving in extreme environments: from genes and genomes to proteins, structures and regulation. Extremophiles. 2021 Nov;25(5-6):425-436.
doi: 10.1007/s00792-021-01242-z
PMID: 34542714
Da Silva GJ, Domingues S. Insights on the horizontal gene transfer of carbapenemase determinants in the opportunistic pathogen Acinetobacter baumannii. Microorganisms. 2016 Aug 23;4(3):29.
doi: 10.3390/microorganisms4030029
PMID: 27681923
Domingues S, Rosário N, Cândido Â, Neto D, Nielsen KM, Da Silva GJ. Competence for natural transformation is common among clinical strains of resistant Acinetobacter spp. Microorganisms. 2019 Jan 24;7(2):30.
doi: 10.3390/microorganisms7020030
PMID: 30682786
Fondi M, Bacci G, Brilli M, Papaleo MC, Mengoni A, Vaneechoutte M, Dijkshoorn L, Fani R. Exploring the evolutionary dynamics of plasmids: the Acinetobacter pan-plasmidome. BMC Evol Biol. 2010 Feb 24;10:59.
PMID: 20181243
Godeux AS, Lupo A, Haenni M, Guette-Marquet S, Wilharm G, Laaberki MH, Charpentier X. Fluorescence-based detection of natural transformation in drug-resistant Acinetobacter baumannii. J Bacteriol. 2018 Sep 10;200(19):e00181-18.
doi: 10.1128/JB.00181-18
PMID: 30012729
Godeux AS, Svedholm E, Barreto S, Potron A, Venner S, Charpentier X, Laaberki MH. Interbacterial transfer of carbapenem resistance and large antibiotic resistance islands by natural transformation in pathogenic Acinetobacter. mBio. 2022 Feb 22;13(1):e0263121.
PMID: 35073754
Erratum in: mBio. 2022 Apr 26;13(2):e0052622. doi: 10.1128/mbio.00526-22
Hu Y, He L, Tao X, Meng F, Zhang J. High DNA uptake capacity of International Clone II Acinetobacter baumannii detected by a novel planktonic natural transformation assay. Front Microbiol. 2019 Sep 24;10:2165.
PMID: 31616393
Hu Y, Zheng J, Zhang J. Natural transformation in Acinetobacter baumannii W068: A genetic analysis reveals the involvements of the CRP, XcpV, XcpW, TsaP, and TonB2. Front Microbiol. 2022 Jan 20;12:738034.
doi: 10.3389/fmicb.2021.738034
PMID: 35126321
Martinez J, Liu C, Rodman N, Fernandez JS, Barberis C, Sieira R, Perez F, Bonomo RA, Ramirez MS. Human fluids alter DNA-acquisition in Acinetobacter baumannii. Diagn Microbiol Infect Dis. 2019 Mar;93(3):183-187.
doi: 10.1016/j.diagmicrobio.2018.10.010
PMID: 30420211
McInnes RS, McCallum GE, Lamberte LE, van Schaik W. Horizontal transfer of antibiotic resistance genes in the human gut microbiome. Curr Opin Microbiol. 2020 Feb;53:35-43.
doi: 10.1016/j.mib.2020.02.002
PMID: 32143027
Quinn B, Martinez J, Liu C, Nguyen M, Ramirez MS. The effect of sub-inhibitory concentrations of antibiotics on natural transformation in Acinetobacter baumannii. Int J Antimicrob Agents. 2018 May;51(5):809-810.
doi: 10.1016/j.ijantimicag.2018.01.026
PMID: 29408470
Ramirez MS, Don M, Merkier AK, Bistué AJ, Zorreguieta A, Centrón D, Tolmasky ME. Naturally competent Acinetobacter baumannii clinical isolate as a convenient model for genetic studies. J Clin Microbiol. 2010 Apr;48(4):1488-90.
doi: 10.1128/JCM.01264-09
PMID: 20181905
Traglia GM, Quinn B, Schramm ST, Soler-Bistue A, Ramirez MS. Serum albumin and Ca2+ are natural competence inducers in the human pathogen Acinetobacter baumannii. Antimicrob Agents Chemother. 2016 Jul 22;60(8):4920-9.
doi: 10.1128/AAC.00529-16
PMID: 27270286
Vesel N, Blokesch M. Pilus production in Acinetobacter baumannii is growth phase dependent and essential for natural transformation. J Bacteriol. 2021 Mar 23;203(8):e00034-21.
doi: 10.1128/JB.00034-21
PMID: 33495250
Wilharm G, Piesker J, Laue M, Skiebe E. DNA uptake by the nosocomial pathogen Acinetobacter baumannii occurs during movement along wet surfaces. J Bacteriol. 2013 Sep;195(18):4146-53.
doi: 10.1128/JB.00754-13
PMID: 23852865